© 2023 IIETA. This article is published by IIETA and is licensed under the CC BY 4.0 license (http://creativecommons.org/licenses/by/4.0/).
OPEN ACCESS
Accurate diagnosis is a crucial first step in the management and treatment of lung diseases, which include infectious diseases such as COVID-19, viral pneumonia, lung opacity, tuberculosis, and bacterial pneumonia. Despite these conditions sharing similar manifestations in chest X-ray images, it is imperative to correctly identify the disease present. This study, therefore, sought to develop a convolutional neural network (CNN)-based model for the classification of lung diseases. Four distinct CNN models, namely MobileNetV2, ResNet-50, ResNet-101, and AlexNet, were rigorously evaluated for their ability to classify lung diseases from chest X-ray images. These models were tested against three classification schemes to examine the impact of high interclass similarity: a 4-subclass classification (COVID-19, viral pneumonia, lung opacity, and normal), a 5-subclass classification (COVID-19, viral pneumonia, lung opacity, tuberculosis, and normal), and a 6-subclass classification (COVID-19, lung opacity, viral pneumonia, tuberculosis, bacterial pneumonia, and normal). The retrained ResNet-50 architecture yielded the best results, achieving a classification accuracy of 97.22%, 92.14%, and 96.08% for the 6-subclass, 5-subclass, and 4-subclass classifications respectively. Conversely, ResNet-101 demonstrated the lowest classification accuracy for the 6-subclass and 5-subclass classifications, with 78.12% and 79.49% respectively, while MobileNetV2 had the lowest accuracy for the 4-subclass classification, with 88.89%. These results suggest that, despite high interclass similarity, the ResNet-50 model can effectively classify lung-related diseases from chest X-ray images. This finding supports the use of computer-aided detection (CAD) systems as decision-support tools in the early classification of lung-related diseases.
deep learning, diagnosis, lung disease, MobileNetV2, ResNet-50, transfer learning
Lung or pulmonary diseases constitute a spectrum of pathological disorders that adversely affect the organs and tissues instrumental in respiration, thereby impeding efficient gas exchange. These afflictions compromise various components of the respiratory system, including the pleurae, the pleural cavity, respiratory nerves, breathing muscles, bronchi, bronchioles, alveoli, and trachea [1]. The spectrum of lung diseases spans minor, self-limiting conditions such as the common cold, influenza, and pharyngitis, to life-threatening diseases like bacterial pneumonia, lung opacity, tuberculosis, acute asthma, lung cancer, and severe acute respiratory syndromes such as COVID-19 [2].
The burden of lung diseases is particularly significant in developing and low- to middle-income countries, where millions grapple with extreme poverty and poor air quality. As estimated by the World Health Organization, over four million individuals succumb prematurely each year to illnesses associated with lung diseases such as asthma and pneumonia. This underscores the urgency for effective preventive measures and diagnostic systems for early detection and management of lung diseases [3, 4].
Since the outbreak in late 2019, COVID-19 has exhibited severe pulmonary implications, including difficulty in breathing among those infected. Moreover, the causative agent of COVID-19, along with other viruses or bacteria, can precipitate pneumonia, a form of lung disease.
The integration of radiological imaging procedures and computer-aided diagnosis (CAD) systems holds great promise for improving the detection and classification of lung diseases. Given that these diseases primarily affect the lung passages, the application of deep learning modalities for early detection and classification is particularly apt. The use of machine vision, image-processing techniques, and deep learning algorithms as diagnostic tools can significantly reduce the risk of misdiagnosis [5]. These tools offer superior accuracy, portability, and affordability, thereby enhancing their applicability in clinical settings. This can substantially mitigate lung-related health risks, especially in regions where access to quality healthcare is limited. The deployment of CAD systems can potentially improve the quality of healthcare services, decreasing mortality rates amidst increasing disease prevalence.
The concept of transfer learning in image classification posits that a model trained on a sufficiently large and diverse dataset can serve as a generalized model of the visual world. Once the feature maps are learned, they can be used to construct a different model using a significant dataset, eliminating the need to start from scratch. Transfer learning involves leveraging knowledge gained from one model to improve the efficacy of another model [6, 7].
Accordingly, in this study, deep learning techniques employing transfer learning were utilized to classify lung diseases. Chest X-ray images were fed into deep learning architectures (MobileNetV2, ResNet-50, ResNet-101, and AlexNet) to identify and extract the pertinent features of these diseases. The objectives of this study were to curate hybrid chest X-ray image datasets of lung diseases from benchmark datasets, preprocess these images to address inconsistencies in data formats, develop a CNN-based model for classifying lung diseases from these chest X-ray images, and evaluate the performance of this classification model.
In their seminal work, Zak and Krzyżak [8] presented a bifurcated deep learning approach for the classification of lung disorders (namely, pneumonia and tuberculosis) based on chest X-ray images extracted from the Shenzhen dataset. The initial phase involved the utilization of U-Net to segment the target lung region, followed by the employment of three transfer learning models (InceptionV3, VGG16, and ResNet-50) with previously trained ImageNet weights. Notably, the InceptionV3 model exhibited superior performance with an accuracy and specificity of 82.00% and a sensitivity of 82.33%.
Similarly, Apostolopoulos and Mpesiana [9] employed transfer learning for classifying lung disorders (normal, COVID-19, and pneumonia) using chest X-ray images from a publicly available dataset. They used five models (Inception, MobileNetV2, Xception, Inception ResNetV2, and VGG19), and VGG19 emerged as the most effective with a specificity of 98.75%, accuracy of 93.48%, and sensitivity of 92.85%.
In an innovative approach, Pham [10] combined three databases, namely the COVID-19 Chest X-Ray Dataset Initiative, the COVID-19 Radiography Database, and the IEEE 8023/COVID Chest X-Ray dataset, for conducting 2-class (COVID-19 and normal cases) and 3-class (COVID-19, viral pneumonia, and normal) classifications. Three pre-trained CNNs, AlexNet, SqueezeNet, and GoogleNet, were incorporated into the network. The model achieved an accuracy of 95%, specificity of 97%, and sensitivity of 90% in detecting COVID-19. Notably, among the applied pre-trained CNNs, SqueezeNet and AlexNet demanded the least training and prediction time.
Further, an end-to-end learning method was proposed by Kim et al. [11], wherein raw chest X-ray images were directly input into a deep learning model (EfficientNet v2-M) using transfer learning to extract significant features for lung disease classification. The method was validated using three classes of data from the National Institutes of Health (NIH) dataset: normal, pneumonia, and pneumothorax. The validation results demonstrated a loss of 0.6933, accuracy of 82.15%, sensitivity of 81.40%, and specificity of 91.65%. The method was further tested on a dataset from the Cheonan Soonchunhyang University Hospital (SCH), which comprised four classes: normal, pneumothorax, tuberculosis, and pneumonia. The testing accuracy for the normal, pneumonia, pneumothorax, and tuberculosis classes was reported as 63.60%, 82.30%, 82.80%, and 89.90%, respectively.
In light of the aforementioned studies, the present study seeks to make the following contributions:
(1) The curation of a hybrid chest X-ray image dataset encompassing COVID-19, bacterial pneumonia, viral pneumonia, lung opacity, tuberculosis, and healthy instances; and
(2) The development of a CNN-based multi-classification model for lung-related diseases using chest X-ray images from multiple sources to enhance the analysis of chest radiography images.
The dataset that was used for this study were gotten from different publicly available online repository. The coronavirus Radiography Database by Chowdhury et al. [12] from Kaggle [13] were accessed and collated in order to fulfill the requirements of this investigation. 3,616 X-rays of chest images belong to the COVID-19 class, 1,345 images of chest X-rays that show viral pneumonia, 6,012 X-rays images of lung opacity, and 10,192 chest X-rays images that are normal in this dataset. From this repository, 1,000 images were put to use from each class of images.
Chest X-Ray for Tuberculosis is the second dataset. CXR images for Tuberculosis (TB) positive cases as well as normal images are included in the dataset from the Kaggle repository that belongs to investigators from Qatar University in Doha, Qatar, and the University of Dhaka, Bangladesh, as well as their collaborators from Malaysia and in collaboration with medical professionals from Hamad Medical Corporation and Bangladesh. This chest X-ray data is available in the study [14]. The dataset includes 700 X-rays of chest images that were gotten for the tuberculosis class. Another tuberculosis data was obtained from the TBX11K dataset [15] which consists of five categories of tuberculosis diseases in this dataset, i.e., Healthy, Sick but Non-TB, Latent TB, Active TB, and Uncertain TB, active TB dataset were extracted for this research. The last dataset that was used for the research which is bacterial pneumonia was also obtained from the Kaggle repository [16].
The total dataset contains 6,000 datasets which were divided into 6 classes. Each class in the dataset contains 1,000 data samples. The data samples in each class were split in the proportion of 80:20 to train and test, while 10 percent of the training samples were used for validation to prevent overfitting. Table 1 illustrates the distribution of the dataset. Figure 1 depicts the images of chest X-rays of viral pneumonia, COVID-19, normal, tuberculosis, lung opacity, and bacterial pneumonia.
Figure 1. Included in the sample images of chest X-ray used in this study (a) COVID 19; (b) Pneumonia; (c) Normal; (d) Tuberculosis positive; (e) lung-opacity; (f) bacterial pneumonia
Table 1. Distribution of the dataset
|
Class |
Total Number of CXIs/Class |
Training Set |
Validation Set |
Testing |
|
COVID-19 |
1000 |
720 |
80 |
200 |
|
Viral Pneumonia |
1000 |
720 |
80 |
200 |
|
Tuberculosis |
1000 |
720 |
80 |
200 |
|
Lung Opacity |
1000 |
720 |
80 |
200 |
|
Bacterial Pneumonia |
1000 |
720 |
80 |
200 |
|
Normal |
1000 |
720 |
80 |
200 |
The normalize function from the Python preprocessing library was used to normalize the data as part of the preprocessing of the dataset. It accepts an array as input and normalizes its values to range from 0 to 1. After that, it gives back an output array with the same size as the input array. Normalization is the process of rescaling attributes of real-valued images into a 0 to 1 range. To lessen the sensitivity of model training to feature scale, deep learning uses data preprocessing. This makes it possible for the model in converging to improved weights, which leads to a more accurate model. Also, the dataset was in different sizes, therefore, they were pre-processed to the same size of 224×224.
The study was implemented with a transfer learning technique by manipulating MobileNetv2, ResNet50, ResNet101, and AlexNet as a base model and defined the new top layers as a fine-tuning model. The conventional pipeline of transfer learning is initially to excerpt features from the source dataset and then fine-tune them on the targeted dataset. Figure 2 shows the transfer learning pipeline.
Figure 2. The transfer learning pipeline
Four cutting-edge pretrained networks—MobileNetv2, ResNet50, ResNet101, and AlexNet—were used in this study in extracting the deep features from the previously trained networks. These networks were refined using the chest x-ray dataset after being pre-trained on the ImageNet dataset, containing one million images divided into 1,000 classes. In this study, the chest X-ray images will be fed into each pre-trained network individually in extracting the highlighted vectors at the fully connected layer. All training was done using the Python programming language in Jupyter Notebook. MobileNetv2 and ResNet50 was implemented with TensorFlow Keras library while ResNet101 and AlexNet were implemented on PyTorch library on Intel(R) Core (TM) i5-2,520M central processing unit operating at 2.50 GHz, and RAM comprised of 8GB 2,400 MHz DDR4 modules.
The model was trained for 6-subclass classification (Coronavirus, tuberculosis, viral pneumonia, bacterial lung opacity, and pneumonia) and 5-subclass classification (COVID-19, viral pneumonia, bacterial pneumonia, lung opacity, and tuberculosis) for all the models. The model was trained for 4-subclass classification (COVID-19, Viral Pneumonia, lung opacity, and bacterial pneumonia) for MobileNetV2 and ResNet-50. Each model has a batch size of 32, drop out of 0.6, a learning rate of 0.001, global average pooling, and Adam optimiser was employed for the precise classification.
The models will be evaluated using five criteria which are accuracy, recall, precision, specificity, and F1-score. They are as described in Eqs. (1)-(4):
$A c c u a c y=\frac{T P+T N}{T P+T N+F P+F N}$ (1)
Precision $=\frac{T P}{T P+F P}$ (2)
Recall $=\frac{T P}{T P+F P}$ (3)
$F 1$ Score $=\frac{2 \,* \text { precision } * \text { Recall }}{\text { Precision }+ \text { Recall }}$ (4)
where, TP (True Positive), TN (True Negative), FN (False Negative) and FP (False Positive).
Also, confusion matrices analysis was used in validating the model [17]. The confusion matrix provides extra details on the illustration of the classification model. The confusion matrix has two types of elements: diagonal and off-diagonal. The diagonal elements depict how many points have predicted labels that match actual labels, indicating that the point is correctly classified. Off-diagonal elements represent how many of the points for which the classifier mislabelled or misclassified the data. The more accurate the predictions that the model was able to make, indicated by the diagonal numbers of the confusion matrix indicates the prediction that the model was able to make accurately.
The model was iterated using the forward propagation as well as the backward propagation utilising Adam optimiser. The optimised repetitions of forward and backward propagation making the model optimised for classification of the lung diseases.
Adam Optimizer is defined as shown in Eq. (5):
$\Delta w_t=\eta \frac{V_t}{\sqrt{S_t+\in}} * g$ (5)
η: Initial learning rate;
Gt: Gradient at time t along wj;
Vt: Exponential average of squares of gradients along wj;
St: Exponential average of squares of gradients along wj; and
$\in$: Smoothing term.
In this section, the distinct metrics for the classification of lung-related diseases were observed, followed by the accuracy observation. The model was trained in detecting and classifying lung disease-confirmed cases by means of chest X-rays images. The dataset was built with a randomly chosen evenly distributed amount of chest x-rays from the chosen repository to eliminate bias effects. Different dataset was used for each of the training, validation, and testing. In essence, the test dataset has not been known aforehand by the model, this will make the model perform excellently on the new dataset.
For each of the 6-subclass classifications, the implementation for lung-related disease cases was trained and validated. For all of them, the model was trained for 10 epochs with a batch size of 32. The results of the analysis of the 6-subclass lung-related diseases with a balanced dataset with the deep feature extraction pipeline are presented.
Figure 3 shows the trained and validated losses of each of the models, whereas Figure 4 showed the trained and validated accuracy for each of the models. In each of the epochs, ResNet50 had the lowest training loss and also had the highest training accuracy. This implies that the lung diseases were able to train well on the ResNet50 model.
Figure 3. Trained and validated accuracy model for six-subclass classification
Figure 4. Trained and validated loss model for six-subclass classification
Figure 5. Confusion matrix for MobileNetv2 for 6-subclass classification for classification of lung-related diseases
The confusion matrix for the 6-subclass classification of lung-related disease dataset with MobileNetv2 is presented in Figure 5. COVID-19, viral pneumonia, and bacteria pneumonia achieved good classification performance in comparison to the other classes of diseases. The model is able to classify the occurrence of one lung disease. This will enable the patient to quickly take adequate precautions. Furthermore, classification measures for each class in terms of precision, recall, specificity, F1-score were also determined (Table 2). The highest precision was for COVID-19, viral pneumonia, and bacterial pneumonia, which demonstrates the method's potential for use in assisting with the quick classification of bacterial, viral, and COVID-19 pneumonia. Table 3 displays the ResNet50 results for macro average metrics for 6-subclass classification. Nevertheless, Table 4 shows the MobileNetv2 results for macro average metrics for 5-subclass classification.
In addition, Figures 6-8 show the confusion matrix of ResNet50, ResNet101 and AlexNet respectively. The models have a good classification for each of the lung-related diseases.
Table 2. MobileNetv2 results for macro average metrics for 6-subclass classification
|
Dataset |
Precision |
Recall |
F1-Score |
Support |
|
Bacteria Pneumonia |
0.93 |
1.00 |
0.96 |
186 |
|
Lung_Opacity |
0.70 |
0.64 |
0.67 |
222 |
|
Viral Pneumonia |
0.94 |
0.56 |
0.70 |
336 |
|
COVID-19 |
0.99 |
0.78 |
0.87 |
255 |
|
Normal |
0.10 |
0.79 |
0.17 |
24 |
|
Tuberculosis |
0.82 |
0.93 |
0.87 |
177 |
Figure 6. Confusion matrix for ResNet50 for 6-subclass classification for the classification of lung-related diseases
Table 3. ResNet50 results for macro average metrics for 6-subclass classification
|
Dataset |
Precision |
Recall |
F1-Score |
Support |
|
Bacteria Pneumonia |
0.94 |
0.99 |
0.97 |
191 |
|
Lung_Opacity |
0.89 |
0.73 |
0.80 |
243 |
|
Viral Pneumonia |
0.94 |
0.65 |
0.77 |
286 |
|
COVID-19 |
0.91 |
0.89 |
0.90 |
205 |
|
Normal |
0.20 |
0.95 |
0.33 |
42 |
|
Tuberculosis |
0.94 |
0.81 |
0.87 |
233 |
Table 4. MobileNetv2 results for macro average metrics for 5-subclass classification
|
Dataset |
Precision |
Recall |
F1-Score |
Support |
|
Lung-Opacity |
0.70 |
0.86 |
0.77 |
163 |
|
Viral Pneumonia |
0.94 |
0.75 |
0.83 |
251 |
|
COVID |
0.99 |
0.84 |
0.91 |
235 |
|
Normal |
0.55 |
0.91 |
0.69 |
121 |
|
Tuberculosis |
0.76 |
0.62 |
0.68 |
170 |
Figure 7. Confusion matrix for ResNet101 for 6-subclass classification for classification of lung-related diseases
Figure 8. Confusion matrix for AlexNet for 6-subclass classification for detection of lung-related diseases
Moreover, MobileNetV2, ResNet50, ResNet101, and AlexNet architectures were trained on a total of 5,000 datasets for five-subclass classification. Chest x-ray consists of 1,000 images each for the lung-related disease (Lung-Opacity, COVID-19, Viral pneumonia, normal, and tuberculosis) with the dataset divided in the proportion of 80:20 for training and testing, the training dataset was additionally divided into training and validation.
Figure 9 illustrates the trained and validated loss of each of the model, whereas Figure 10 shows the trained and validated accuracy for each of the model. In each of the epochs, ResNet50 had lowest training loss and had the highest training accuracy. This implies that the lung diseases were able to train well on the ResNet50 model.
Figure 11 depicts Confusion matrix for MobileNetv2 for 5-subclass classification for the classification of lung-related diseases while Figure 12 shows the confusion matrix for ResNet101 for 5-subclass classification for detection of lung-related diseases while Figures 12 and 13 show the confusion matrix for ResNet50 and ResNet101 for 5-subclass classification for detection of lung-related diseases respectively. Finally, Figure 14 shows the confusion matrix for AlexNet for 5-subclass classification for the classification of lung-related diseases.
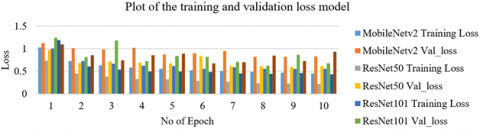
Figure 9. Trained and validated loss model for five-subclass classification
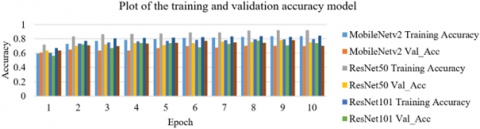
Figure 10. Trained and validated accuracy model for five-subclass classification
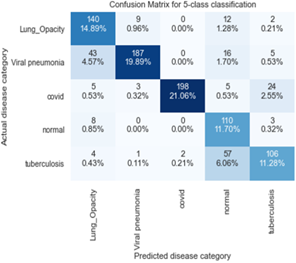
Figure 11. Confusion matrix for MobileNetv2 for 5-subclass classification for the classification of lung-related diseases

Figure 12. Confusion matrix for ResNet101 for 5-subclass classification for detection of lung-related diseases
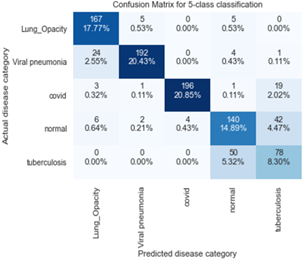
Figure 13. Confusion matrix for ResNet50 for 5-subclass classification for detection of lung-related diseases
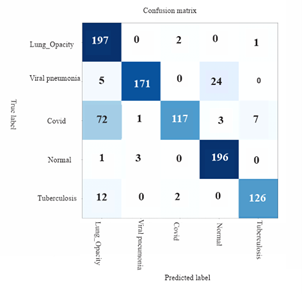
Figure 14. Confusion matrix for AlexNet for 5-subclass classification for the classification of lung-related diseases
In this study, MobileNetV2 and ResNet50 were built as lightweight deep-learning models for lung-related disease identification and classification in chest X-rays. Also, ResNet101 and AlexNet models were also trained using the same dataset with PyTorch libraries. The architecture of the model is used in the classification of the most prevalent and pandemic lung-related diseases: bacterial pneumonia, lung opacity, viral pneumonia, COVID, normal, and tuberculosis. The model compared the performance of the accuracy, recall, precision, F1 score. The highest training accuracy was achieved 97% and the testing accuracy is 94% indicating that model is able to classify each of the lung-related diseases accurately.
For six classifications, ResNet-50 outperformed MobileNetV2 in terms of accuracy, as evidenced by the confusion matrices and average accuracies. In ten (10) epochs, the accuracy of the ResNet-50 is 97%, whereas that of the MobileNetV2 is 86.35%. However, MobileNet's training has the lowest time. In the process of increasing the number of epochs, accuracy will undoubtedly continue to rise. In comparison to MobileNet, ResNet-50, ResNet101, and AlexNet, ResNet50 has the highest training and testing accuracy for the detection of lung-related diseases. The accuracy of each of the model is shown in Table 5. Furthermore, Table 6 shown the comparison of the training time for each of the model and classification.
Table 5. Comparison with earlier literature on classification of lung-related diseases with X-ray images of the chest
|
Author(s) |
Dataset |
Techniques Used |
Accuracy |
|
Al-Timemy et al. [18] |
5-subclasses: 435 COVID-19/ 439 normal/ 439 pneumonia bacterial/ 434 Tuberculosis/ 439 pneumonia viral 3-subclasses: 435 COVID-19/ 434 Tuberculosis / 439 normal |
The subspace discriminant classifier ensemble of Resnet50 |
91.6 98.6 |
|
Jia et al. [19] |
5-subclasses 1770 COVID-19, 1436 Tuberculosis, 1345 viral pneumonia, 1700 bacterial pneumonia and 1341 normal 4-subclasses 1700 COVID-19, 1341, viral pneumonia, 1436 tuberculosis, 1341 healthy 3-subclasses 1770 COVID-19, 1341 viral pneumonia, and 1341 healthy |
Modified MobileNet |
99.6 99.9 99.7 |
|
Ozturk et al. [20] |
3-subclass: 125 COVID-19/ 500 Pneumonia/ 500 Normal |
DarkCOVIDNet CNN |
87.2 |
|
Khan et al. [21] |
4-subclass: 284 COVID-19/ 310 normal/ 330 pneumonia bacterial/ 327 pneumonia viral |
CoroNet CNN |
89.6 |
|
This study |
6 subclasses 1000 Bacteria pneumonia, 1000 Lung-Opacity, 1000 Viral pneumonia, 1000 COVID, 1000 normal, 1000 tuberculosis |
MobileNetV2 ResNet50 ResNet101 AlexNet |
92 97 82 85 |
|
|
5 subclasses 1000 Lung-Opacity, 1000 Viral pneumonia, 1000 COVID, 1000 normal, 1000 tuberculosis 4 subclasses |
MobileNetV2 ResNet50 ResNet101 AlexNet |
84 91 79 84 |
|
|
1000 Lung-Opacity, 1000 Viral pneumonia, 1000 COVID, 1000 normal |
MobileNetV2 ResNet50 |
88 96 |
Table 6. Comparison of the training time
|
Model |
Classification |
Time (Sec) |
|
ResNet50 |
6- Subclass |
190.18 |
|
MobileNetV2 |
6 - Subclass |
71 |
|
ResNet50 |
5- Subclass |
150.56 |
|
MobileNetV2 |
5 - Subclass |
40.86 |
|
ResNet50 |
4- Subclass |
189.53 |
|
MobileNetV2 |
4 - Subclass |
34.54 |
In the present study, a multi-pronged approach was adopted, leveraging the capabilities of MobileNetv2, ResNet50, ResNet101, and AlexNet models. These models, endowed with the previously trained weights of ImageNet and fine-tuned, were deployed for the categorization of lung-related diseases based on chest X-ray images. The overarching aim was to augment the diagnostic capabilities of Computer-Aided Diagnostics (CADs), particularly in terms of accuracy and efficiency.
Lung disorders exhibit chest X-ray characteristics that bear a striking similarity to each other, rendering early classification a matter of paramount importance. In this context, the present study's focus on the utilization of pre-trained networks, devoid of GPU support, emerges as a key feature. A CPU-enabled computer was harnessed for the efficient extraction of deep features from chest X-ray images.
Moreover, the classification accuracy was evaluated across six, five, and four subclass classifications. This study demonstrated the potential of pipelines requiring minimal computational power, thereby contributing to the classification of lung-related diseases using chest X-ray images, particularly in scenarios where more sophisticated or computationally intensive techniques might be unavailable.
The authors acknowledge the part sponsorship of the Covenant University Centre for Research, Innovation, and Discovery (CUCRID), Ota, Ogun State, Nigeria.
[1] Veale, D. (2006). Chronic respiratory care and rehabilitation in France. Chronic Respiratory Disease, 3(4): 215-216. https://doi.org/10.1177/1479972306070070
[2] Yimer, F., Tessema, A.W., Simegn, G.L. (2021). Multiple lung diseases classification from chest X-ray images using deep learning approach. International Journal of Advanced Trends in Computer Science and Engineering, 10(5): 2936-2946. https://doi.org/10.30534/ijatcse/2021/021052021
[3] Mondal, M.R.H., Bharati, S., Podder, P., Podder, P. (2020). Data analytics for novel coronavirus disease. Informatics in Medicine Unlocked, 20: 100374. https://doi.org/10.1016/j.imu.2020.100374
[4] Adetiba, E., Abolarinwa, J.A., Adegoke, A.A., Taiwo, T.B., Ajayi, O.T., Abayomi, A., Badejo, J.A. (2022). DeepCOVID-19: A model for identification of COVID-19 virus sequences with genomic signal processing and deep learning. Cogent Engineering, 9(1): 2017580. https://doi.org/10.1080/23311916.2021.2017580
[5] Xie, Y., Wu, Z., Han, X., Wang, H., Wu, Y., Cui, L., Chen, Z. (2020). Computer-aided system for the detection of multicategory pulmonary tuberculosis in radiographs. Journal of Healthcare Engineering. https://doi.org/10.1155/2020/9205082
[6] Okokpujie, K., John, S., Ndujiuba, C., Badejo, J.A., Noma-Osaghae, E. (2021). An improved age invariant face recognition using data augmentation. Bulletin of Electrical Engineering and Informatics, 10(1): 179-191. https://doi.org/10.11591/eei.v10i1.2356
[7] Okokpujie, K., John, S., Ndujiuba, C., Noma-Osaghae, E. (2020). Development of an adaptive trait-aging invariant face recognition system using convolutional neural networks. In Information Science and Applications: ICISA 2019, Singapore, pp. 411-420. https://doi.org/10.1007/978-981-15-1465-4_41
[8] Zak, M., Krzyżak, A. (2020). Classification of lung diseases using deep learning models. In International Conference on Computational Science, Amsterdam, the Netherlands, pp. 621-634. https://doi.org/10.1007/978-3-030-50420-5_47
[9] Apostolopoulos, I.D., Mpesiana, T.A. (2020). COVID-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, 43: 635-640. https://doi.org/10.1007/S13246-020-00865-4
[10] Pham, T.D. (2021). Classification of COVID-19 chest X-rays with deep learning: New models or fine tuning? Health Information Science and Systems, 9: 1-11. https://doi.org/10.1007/s13755-020-00135-3
[11] Kim, S., Rim, B., Choi, S., Lee, A., Min, S., Hong, M. (2022). Deep learning in multi-class lung diseases’ classification on chest X-ray images. Diagnostics, 12(4): 915. https://doi.org/10.3390/DIAGNOSTICS12040915.
[12] Chowdhury, M.E., Rahman, T., Khandakar, A., Mazhar, R., Kadir, M.A., Mahbub, Z.B., Islam, M.T. (2020). Can AI help in screening viral and COVID-19 pneumonia? IEEE Access, 8: 132665-132676. https://doi.org/10.1109/ACCESS.2020.3010287
[13] COVID-19 radiography database. https://www.kaggle.com/tawsifurrahman/COVID19-radiography-database.
[14] Rahman, T., Khandakar, A., Kadir, M.A., Islam, K.R., Islam, K.F., Mazhar, R., Chowdhury, M.E. (2020). Reliable tuberculosis detection using chest X-ray with deep learning, segmentation and visualization. IEEE Access, 8: 191586-191601. https://doi.org/10.1109/ACCESS.2020.3031384
[15] Liu, Y., Wu, Y.H., Ban, Y., Wang, H., Cheng, M.M. (2020). Rethinking computer-aided tuberculosis diagnosis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, pp. 2646-2655. https://doi.org/10.1109/CVPR42600.2020.00272
[16] Kermany, D.S., Goldbaum, M., Cai, W., Valentim, C.C., Liang, H., Baxter, S.L., Zhang, K. (2018). Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell, 172(5): 1122-1131. https://doi.org/10.1016/J.CELL.2018.02.010
[17] Ruuska, S., Hämäläinen, W., Kajava, S., Mughal, M., Matilainen, P., Mononen, J. (2018). Evaluation of the confusion matrix method in the validation of an automated system for measuring feeding behaviour of cattle. Behavioural Processes, 148: 56-62. https://doi.org/10.1016/j.beproc.2018.01.004
[18] Al-Timemy, A.H., Khushaba, R.N., Mosa, Z.M., Escudero, J. (2021). An efficient mixture of deep and machine learning models for COVID-19 and tuberculosis detection using x-ray images in resource limited settings. Artificial Intelligence for COVID-19, 77-100. https://doi.org/10.1007/978-3-030-69744-0_6
[19] Jia, G., Lam, H.K., Xu, Y. (2021). Classification of COVID-19 chest X-Ray and CT images using a type of dynamic CNN modification method. Computers in Biology and Medicine, 134: 104425. https://doi.org/10.1016/J.COMPBIOMED.2021.104425
[20] Ozturk, T., Talo, M., Yildirim, E.A., Baloglu, U.B., Yildirim, O., Acharya, U.R. (2020). Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine, 121: 103792. https://doi.org/10.1016/j.compbiomed.2020.103792
[21] Khan, A.I., Shah, J.L., Bhat, M.M. (2020). CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Computer Methods and Programs in Biomedicine, 196: 105581. https://doi.org/10.1016/j.cmpb.2020.105581